Our Projects
Precision Medicine integrating Lipidomics
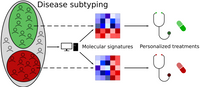
Molecular disease subtying and patient stratification
Precision medicine requires tools for molecular patient stratification to find disease subtypes and develop specialized treatments. We are developing computational tools to stratify patients based on lipidomics or other omics profiles. Our approaches are based on biclustering algorithms, which we modify and extend to improve performance. This allows us to identify molecular signatures that characterize patient subgroups, which can serve as markers to classify patients or used as hypotheses, to further investigate the molecular mechanisms of diseases.
Link to preprint: https://www.biorxiv.org/content/10.1101/2021.09.30.462567v1
Link to GitLab repository: https://gitlab.lrz.de/lipitum-projects/mosbi
Link to webtool: https://exbio.wzw.tum.de/mosbi/
LipiTUM team members: Tim Rose, Thibault Bechtler, Octavia Ciora, Florian Molnar, Kim Anh Lilian Le
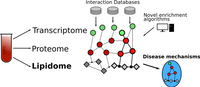
Multi-level omics data is getting available for an increasing number of experiments. This requires computational tools being able to integrate and datamine heterogeneous types of data and produce insights going beyond the separate analysis of each data set. Such tools usually require databases, with functional interactions, such as biological networks. While methods are already available and used in other omics disciplines, they are only partially applicable to lipidomics data. We are utilizing the content of different lipid-protein interaction databases to generate lipid interactions networks. Based on those we develop dedicated algorithms to mechanistically understand lipidome alterations and functionally integrate lipidomics with other omics data.
Link to GitLab repository: https://gitlab.lrz.de/lipitum-projects/linex
Link to webtool: https://exbio.wzw.tum.de/linex/
Link to publication: https://www.mdpi.com/2218-1989/11/8/488
LipiTUM team members: Tim Rose, Nikolai Köhler, Lisa Falk, Lucie Klischat
-----
Deep Learning for Lipid Identification
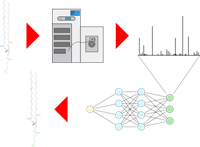
The identification of lipids from fragmented mass spectra at a high structural resolution as well the accurate estimation of False Discovery Rates (FDR) still poses a major challenge in computational lipidomics. In a collaboration with the Analytical BioGeoChemistry research unit at the Helmholtz Center Munich we are aiming at training a neural network based model to accurately identify lipids based on their mass-spectra, by learning the relationships between molecular lipid structures and fragmentation patterns in mass spectra in different fragmentation settings. Having shown the ability of neural networks to learn such relations, it will also be possible to predict lipid spectra more accurately and to obtain better FDR estimations.
LipiTUM team members: Nikolai Köhler, Vivian Würf
-----
Unravelling the influence of microbiota
The activation of activating transcription factor 6 (ATF6) has recently been associated with reduced disease-free survival time in Colorectal Cancer (CRC) patients and was shown to promote colorectal tumorigenesis in a microbiome-dependent fashion in transgenic mice. In this collaboration with the Chair of Nutrition and Immunology we are working on identifying the mechanism behind the tumor-promoting role of ATF6. Our contribution is the development of a novel method for the computational integration of metabolome data with microbiome and transcriptome data and subsequent data and network analysis. The focus of our method lies on allowing the incorporation of prior knowledge and the identification of mechanistic patterns.
LipiTUM team members: Nikolai Köhler, Vivian Würf
We are collaborating with the LiSyM consortium to analyze clinical lipidomics samples from patients with non-alcoholic fatty liver disease and nonalcoholic steatohepatitis. By applying established and our novel machine learning methods, we are seeking to identify disease specific markers in the lipid markers and patient subgroups. Molecular lipidomics data could help to provide early detection of these widespread diseases and allow personalized treatments.
Link to publication: https://www.jlr.org/article/S0022-2275(21)00086-9/fulltext
LipiTUM team members: Tim Rose
In a collaboration with the Max Planck Institute of Molecular Cell Biology and Genetics and the Paul Langerhans Institute in Dresden, we investigate mechanisms and markers for diabetes in different pancreatic tissues. With our computational expertise we extract relevant molecular signatures to unravel disease mechanisms and their interplay with cellular lipidomes.
LipiTUM team members: Tim Rose
-----
HCC and Hepatitis Viruses
In this project with the University Hospital Regensburg we are investigating the lipidome changes in the liver induced by Hepatitis B Virus (HBV) and Hepatitis C Virus (HCV) in Hepatocellular Carcinoma (HCC) tissue and tumor-adjacent tissue of HCC patients. HCC is the most frequent type of liver cancer and is highly associated with chronic infections with HBV and HCV. Our main role in this project is lipidomics data analysis, especially focusing on the identification of patient-group specific lipid signatures.
LipiTUM team members: Nikolai Köhler
-----
Multi-Organ Methodology and HCC/Cachexia
While systemic diseases frequently affect numerous organs of one organism, only few methods for integrating multi-organ data are available. Together with the Molecular Tumor Biology unit from the RWTH University Hospital Aachen and the Shevchenko Lab from the Max-Planck-Institute for Molecular Cellbiology and Genetics we are investigating the systemic effects of Hepatocellular Carcinoma (HCC) and associated Cachexia by analysing the changes in lipid composition in different (distant) organs. By working towards advanced computational methods for multi-organ integration, we are aiming at improving the understanding of these diseases and providing new tools for studying multi-organ data.
LipiTUM team members: Nikolai Köhler
-----
CoVex: The Corona Virus Explorer
We participated in the development of the Coronavirus Explorer (CoVex), a systems medicine platform for the analysis of host-virus interactions. With the integration of multiple databases and novel algorithms, new drug targets and drug repurposing candidates can be predicted using experimentally validated interactions of the host and viral proteins. This can help to get a better understanding of the viral mechanisms and assist in the search for effective treatments.
Link to publication: https://www.nature.com/articles/s41467-020-17189-2
Link to review: https://www.nature.com/articles/s43588-020-00007-6
LipiTUM team members: Tim Rose
Selected Publications
1. Lipid imaging: Ellis SR et al. Nature Methods (2018). in press.
2. Lipidomics standardization: Pauling JK et al. PLOS ONE. (2017) 12(11): e0188394.
3. Quantitative lipidomics: Gallego SF et al. Biochimica et Biophysica Acta. (2017) 1862(2):145-155.
4. Computational lipidomics: Pauling JK et al. J. Integr. Bioinform. (2017) 13(1): 34-51.
5. MSn lipid profiling: Almeida R et al. J. Am. Soc. Mass Spectrom. (2015) 26(1):133-148.
6. Multi-omics and systems biology: Pauling JK et al. Integr Biol. (2014) 6(11):1058-1068.
7. Multi-omics and networks: Alcaraz N et al. BMC Syst. Biol. (2014) 8:99.
8. Clinical microbiota analysis: Zakharkina T et al. PLOS ONE (2013) 8(7): e68302.
9. Genome transfer: Pauling JK et al. Nucleic Acids Res. (2012) 40(D1):D610-4.
10. Metabolomics: Hauschild AC et al. Metabolites (2012) 2, 733-755.